How to write a method for the INHECO On Deck Thermal Cycler on firefly+
Setup Temperature Initialize Step (mandatory)
- Create and save a new ODTC Script Editor project. A blank page will show three tabs: a) Temp. Init. Steps; b) Methods; c) PCR.
 To start writing a method (e.g. static incubations profile, PCR thermal profile), a pre-method must be set. To do this, either right-click the Temp. Init. Steps tab and Add new Temperature Initialize Step or select that option from the Add menu.
To start writing a method (e.g. static incubations profile, PCR thermal profile), a pre-method must be set. To do this, either right-click the Temp. Init. Steps tab and Add new Temperature Initialize Step or select that option from the Add menu.
-
In the Temperature Initialization Step Setup window set the parameters as follows:
- Plate Type – select the appropriate plate mount (96 or 384 wells).
- Target temperature Mount [°C] – set the target temperature for the mount. In this example, the mount temperature is set to 25 °C (approx. room temperature).
- Target temperature LID [°C] – if left locked, the default lid temperature will be set to 110 °C. To change this parameter, click the yellow padlock icon and edit the field. In this example, the lid temperature is set to 100 °C.

Setup an incubation profile
- To setup an incubation profile (e.g. ligation reaction), either right-click the Methods tab and add new Method or select that option from the Add menu.

-
In the Method Setup window set the parameters as follows:
-
Plate Type – select the appropriate plate mount (96 or 384 wells).
-
Fluid quantity – set the volume range according to the reaction volume in each well (in the example, for a 50uL ligation reaction the 30-74uL range was selected).
-
Keep temperatures after method (Post Temper) – if this option is left On then the last temperature in the profile will be kept on hold (e.g. 10 °C forever).
-
*** Max cooling/heating slopes** – if this option is left On then the maximum 4.4 k/s slope is applied to each step. Select Off if you want to edit each cooling/heating slope. Once setup is complete, this feature cannot be changed. In this example, this feature was left On.
-
Start temperature Mount [°C] – set the start temperature for the mount. In this example, the mount temperature is set to 25 °C (approx. room temperature).
-
Start temperature LID [°C] – if left locked, the default lid temperature will be set to 110 °C. To change this parameter, click the yellow padlock icon and edit the field. In this example, the lid temperature is set to 100 °C.

-
- In the Step Editor table type in the temperature (Celsius) and duration (seconds) for each incubation step. As one temperature entry is completed, another row is automatically generated. In this example, a ligation (step 1) is performed at 25 °C for 10 minutes (600 s), followed by a heat inactivation (step 2) at 65 °C for 10 minutes, and an indefinite hold temperature (step 3; notice the dashed trace at the end of the thermal profile).
 Setup a PCR profile
Setup a PCR profile
Using the PCR tab
-
To setup a standard PCR profile, either right-click the PCR tab and add new PCR or select that option from the Add menu. Note that this approach is less customizable and suited for streamlined PCR setup. If you need more flexibility (e.g. edit different annealing temperatures, ramps, etc) proceed to Using the Method tab instead.

-
In the PCR Setup window set the parameters as follows:
-
Plate Type – select the appropriate plate mount (96 or 384 wells)
-
Fluid quantity – set the volume range according to the reaction volume in each well (in the example, for a 50uL reaction volume the 30-74uL range was selected).
-
Keep temperatures after method (Post Temper) – if this option is left On then the last temperature in the profile will be kept on hold (e.g. 10 °C forever).
-
Start temperature Mount [°C] – set the start temperature for the mount. In this example, the mount temperature is set to 25 °C (approx. room temperature).

-
-
In the PCR Assistant section select between 3-Step or 2-Step PCR type. In this example, a 10 cycles 3-step PCR (separate annealing and elongation settings) is performed as described: i) Initial Denaturation at 98 °C for 180 seconds; ii) Denaturation at 98 °C for 45 seconds; iii) Annealing at 62 °C for 30 seconds; iv) Elongation at 68 °C for 120 seconds; v) Cycle steps ii-iv ten times; vi) Final Extension at 68 °C for 60 seconds; vii) Cooling at 10 °C for 10 seconds and hold temperature.

Using the method tab
-
To setup a custom PCR profile (e.g. nested), either right-click the Methods tab and add new Method or select that option from the Add menu.
-
In the Method Setup window set the parameters as follows:
-
Plate Type – select the appropriate plate mount (96 or 384 wells).
-
Fluid quantity – set the volume range according to the reaction volume in each well (in the example, for a 50uL reaction volume the 30-74uL range was selected).
-
Keep temperatures after method (Post Temper) – if this option is left On then the last temperature in the profile will be kept on hold (e.g. 10 °C forever).
-
*** Max cooling/heating slopes** – if this option is left On then the maximum slope is applied to each step. Select Off if you want to edit each cooling/heating slope. Once setup is complete, this feature cannot be changed. In this example, the feature was left Off.
-
Start temperature Mount [°C] – set the start temperature for the mount. In this example, the mount temperature is set to 25 °C (aprox. room temperature).
-
Start temperature LID [°C] – if left locked, the default lid temperature will be set to 110 °C. To change this parameter, click the yellow padlock icon and edit the field. In this example, the lid temperature is set to 100 °C.

-
- In the Step Editor table type in the heating/cooling slope (K/s) according to the Maximum Table on the right, the temperature (Celsius) and duration (seconds) for each plateau step plateau. As one temperature entry is completed, another row is automatically generated. To loop steps, follow the template:
Step A
Step B
Step C
Step D (must be equal to step A) Go back to: Step B Loop: x-times
Step E (must be equal to step B)
Step F (must be equal to step C)
Following this template and for a loop equal to 8, the script will execute a total of 10 cycles:
1st cycle, steps A/B/C; 2nd - 9th cycles, loop steps D/B/C; 10th cycle, steps D/E/F).
In the example, a nested PCR is setup as follows:

This profile performs 10 cycles with a 53 °C annealing temperature, followed by 5 cycles with a 60 °C annealing temperature and a final cooling at 10 °C forever.

Export edited script to XML format
-
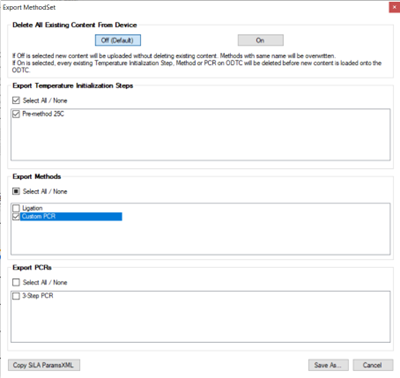
To export an ODTC script in XML format go to the Tools menu and select Export MethodSet (shortcut Ctrl+E).
-
Select which sections of the method need to be exported and Save As.
-
It is recommended that the Temperature Initialization Step and the method be exported in the same XML, since the firefly+ software can use the same script to perform a Pre-Method and Main Method.
For the purpose of this tutorial, the [Pre-method 25C](# Setup-Temperature-Initialize-Step-(mandatory)) and Custom PCR were exported together.
Setup Incubate step on firefly+ using the ODTC script
- In the firefly+ software, add a PCR plate, Lid, and Thermocycler Module assets.
- Add a Place On step to place a Lid on the PCR plate.
- Add a Place On step to move the PCR plate onto the ODTC.
- Add two Incubate steps, one for pre-method ([Temp. Init. Step](# Setup-Temperature-Initialize-Step-(mandatory))) and another to run the main method (Custom PCR in the case of this example).
- In the first Incubate step Properties, set the Method Type to Run Pre method; select the Method File field and embed the ODTC XML script.

- Repeat the previous step for the second Incubate step, selecting Run Main method in the Method Type field.

- Add a Take Off step to retrieve the PCR plate from the ODTC.
- Add a Take Off step to remove the Lid from the top of the PCR plate.

.png?width=300&height=157&name=spt%20logo%20png%20(1).png)